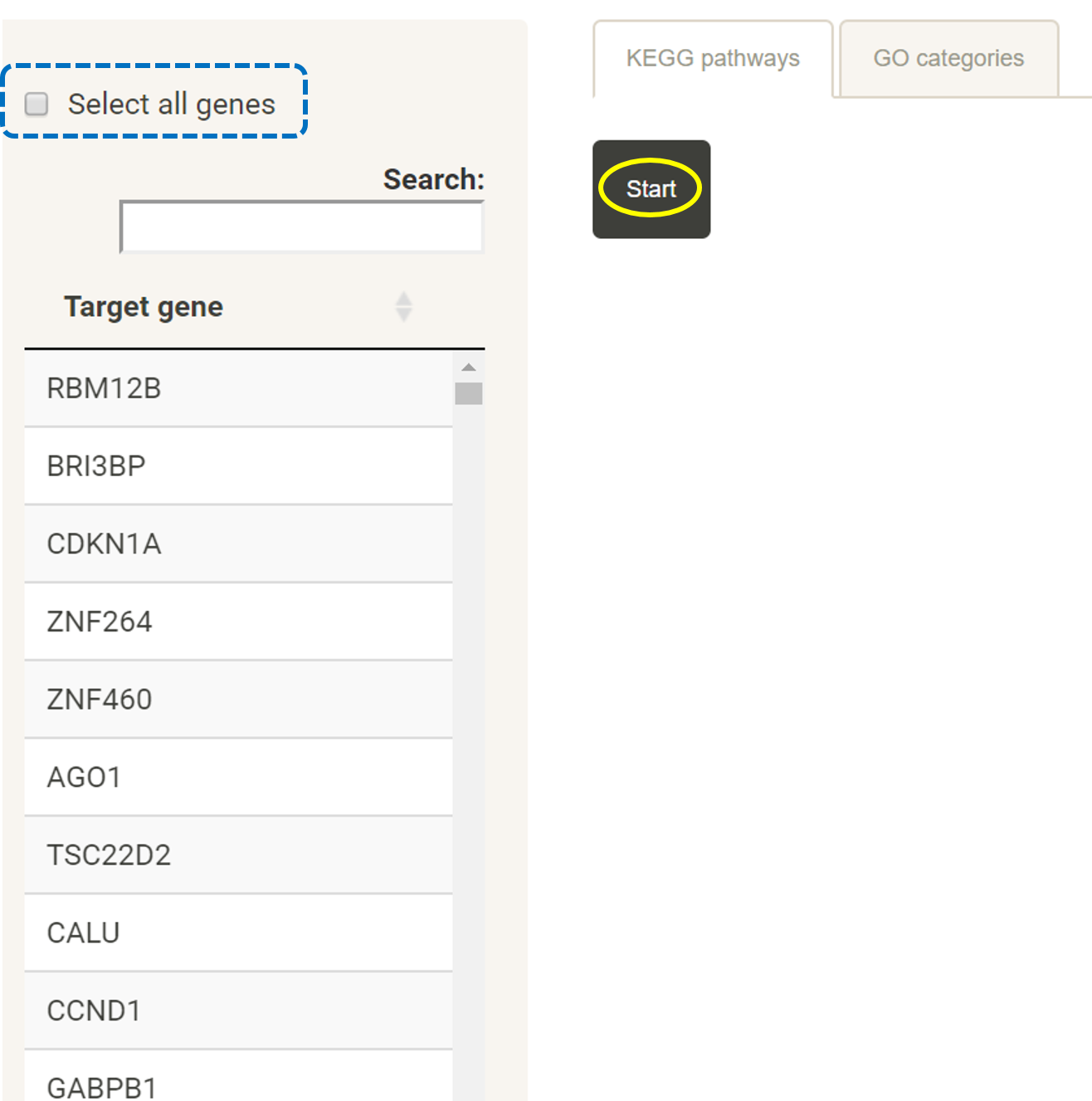
KEGG pathway analysis with target genes for uploaded miRNAs
Prepare a list of genes
After users have uploaded a list of miRNAs, users can first browse the expression levels of these miRNAs and their target genes in different cancer types in TACCO. Users can select a cancer type and the expression levels, fold change and p-values for the uploaded miRANs or their target genes in that cancer type will be listed in the table. Users can click on "Go to pathway analysis".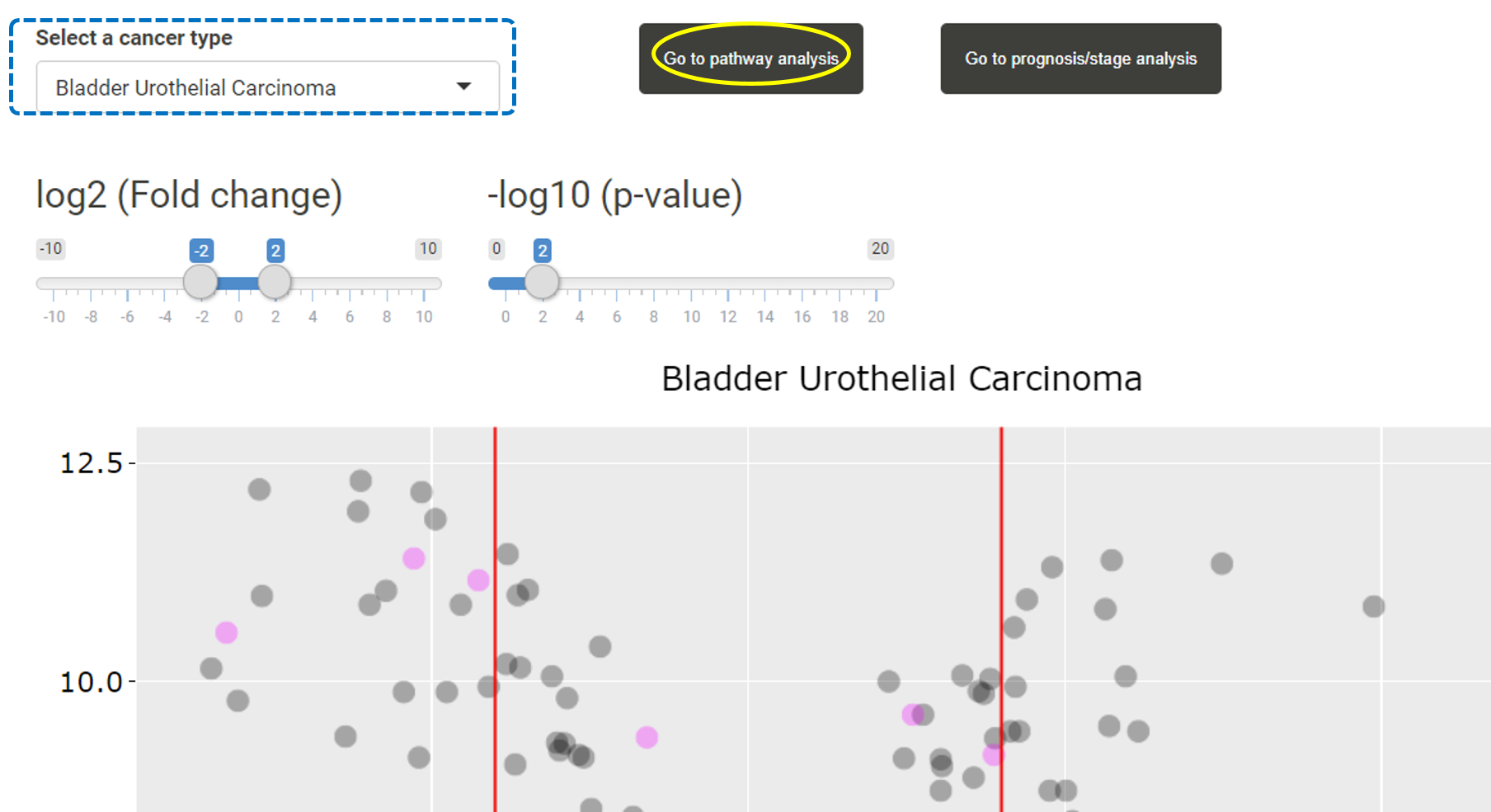
KEGG pathway enrichment results
TACCO provides the gene ratio distribution plot for enriched KEGG pathways. Users can download, zoom in, zoom out or reset the plot.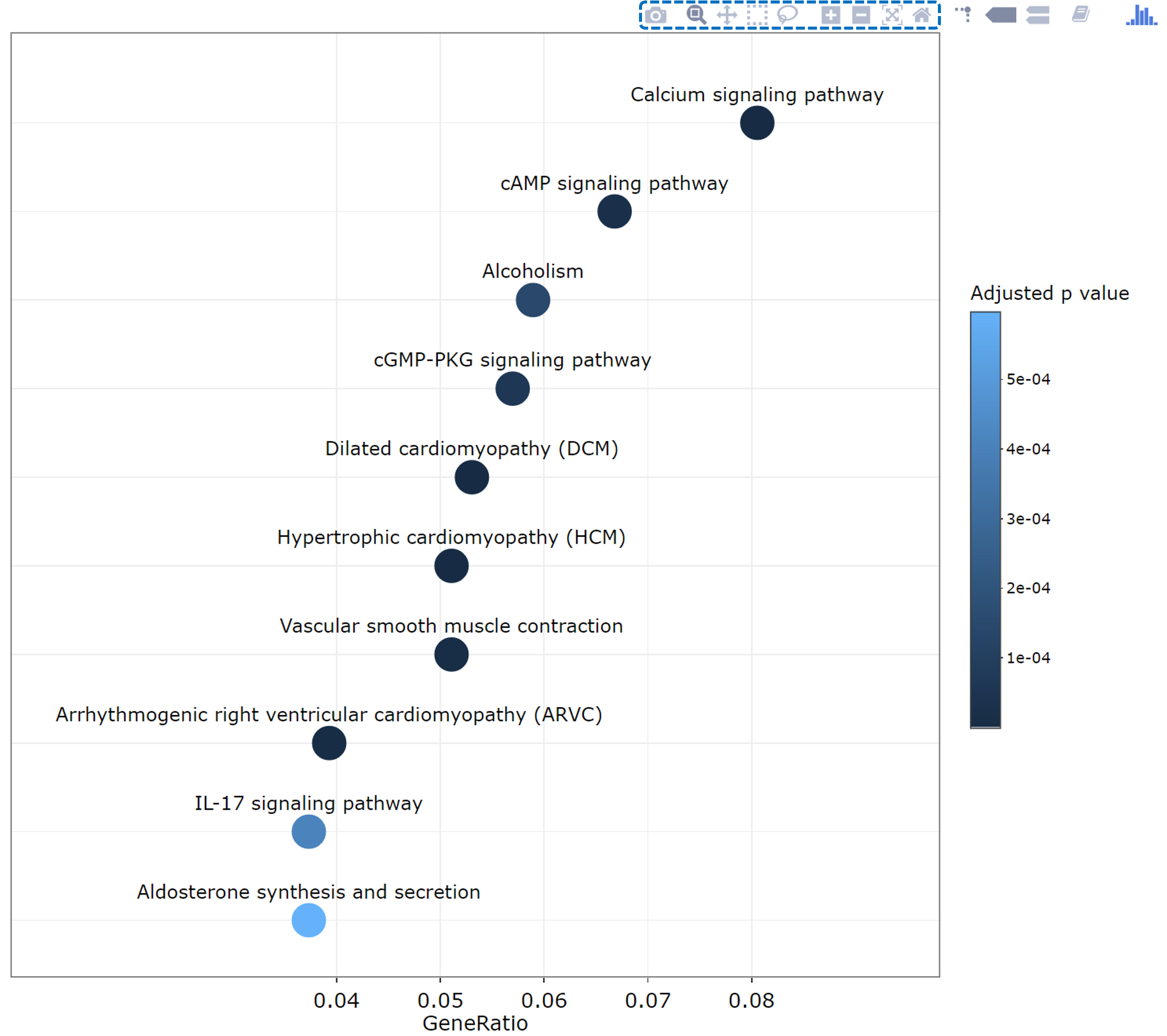
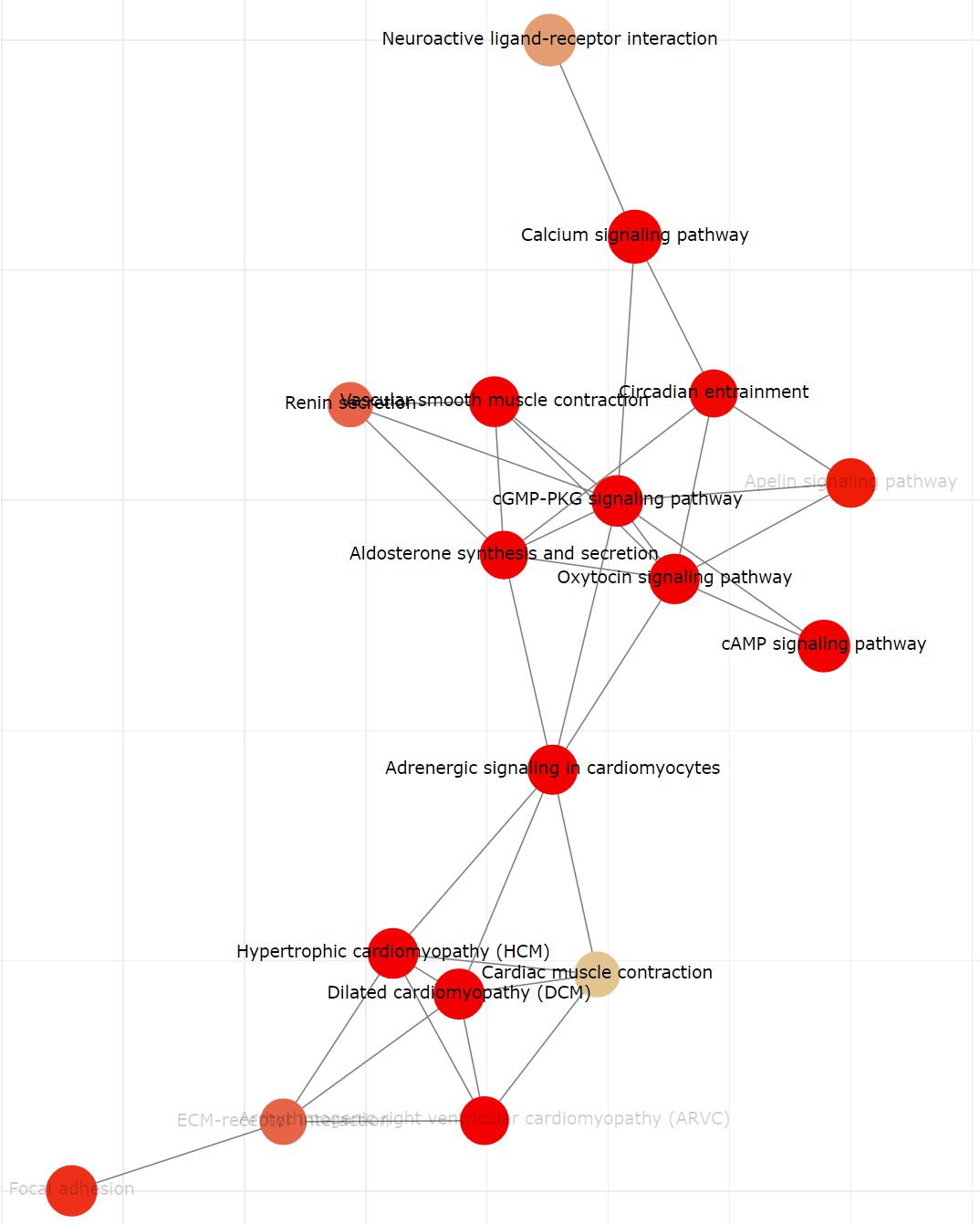
TACCO also provides the connected network plot for enriched KEGG pathways. Pathways sharing the same gene(s) are connected in the network plot. Users can also download, zoom in, zoom out or reset the plot.
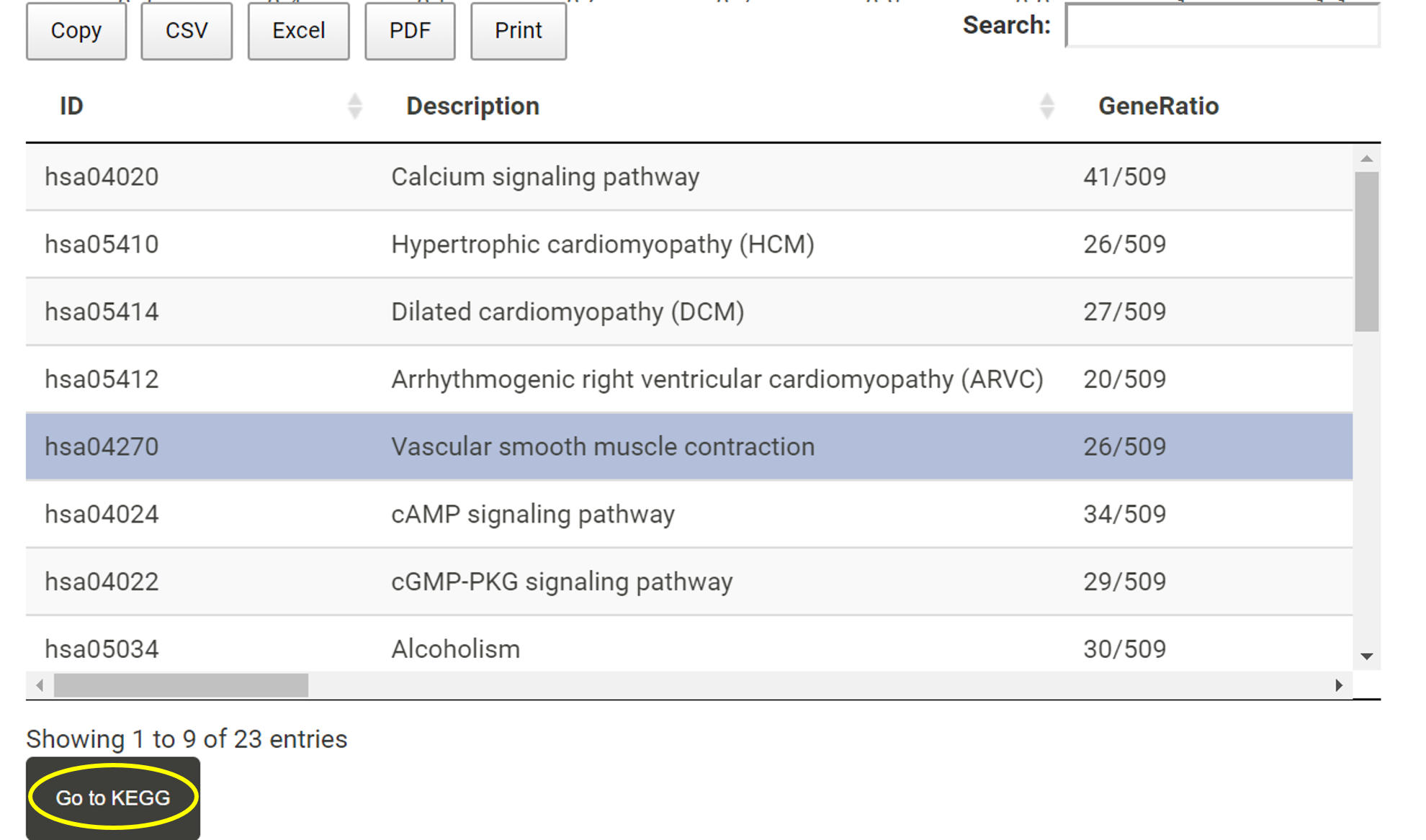
Detailed information for KEGG enrichment analysis is listed in the table. Users can download the table or select a pathway of interest and visit KEGG website. TACCO will open a new page for users to explore the selected pathway in KEGG.
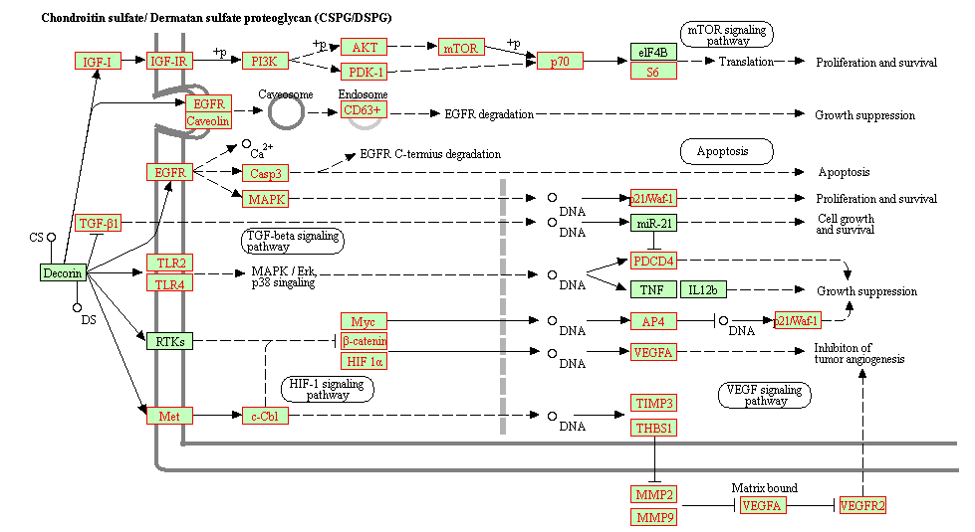
A KEGG pathway example, in which TACCO highlight all the selected genes with the red boxes.